Laboratory #1 - Experimental modeling: model building from experimental data - Prof. M. Taragna
Parametric estimation of position transducer models using the classical (or statistical) approach
Contents
- Introduction
- Procedure
- Problem setup
- Parametric estimation of a linear model (w.r.t. data) using least squares
- Computation of the parameter confidence intervals (noise variance derived from priors)
- Computation of the parameter confidence intervals (noise variance estimated from data)
- Parametric estimation of 3rd order polynomial models (w.r.t. data) using least squares
Introduction
The program code may be splitted in sections using the characters "%%". Each section can run separately with the command "Run Section" (in the Editor toolbar, just to the right of the "Run" button). You can do the same thing by highlighting the code you want to run and by using the button function 9 (F9). This way, you can run only the desired section of your code, saving your time. This script can be considered as a reference example.
clear all, close all, clc
Procedure
- Load the file sensor.mat containing the position/voltage data
- Plot the voltage measurements versus the position measurements
- Estimate the parameters of the linear approximation using least squares
- Plot the estimated approximation versus the experimental data
- Evaluate the estimate uncertaity using the prior information only
- Plot these confidence intervals versus the estimated approximation
- Evaluate the estimate uncertaity using the experimental data only
- Plot these confidence intervals versus the estimated approximation
- Estimate the parameters of polynomial approximations using least squares
- Plot the estimated approximations versus the experimental data
Problem setup
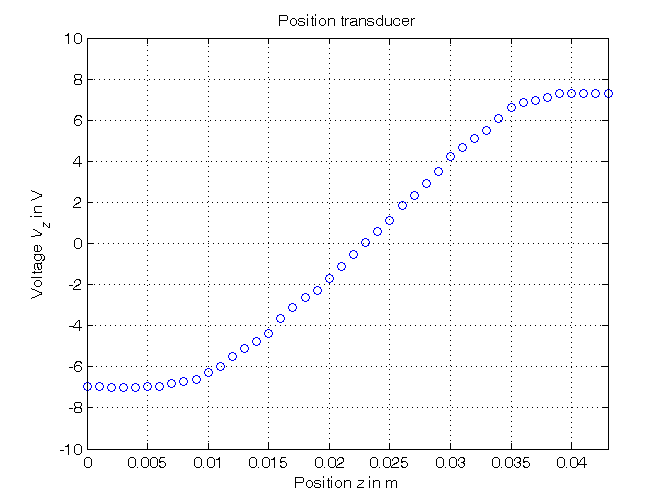
% Step 1: load of data load sensor % z = position measured in meters, without position offset % Vz = voltage measured in volts % Step 2: plot of data figure, plot(z,Vz,'o'), axis([min(z),max(z),-10,10]), grid, title('Position transducer'), xlabel('Position{\it z} in m'), ylabel('Voltage{\it V_z} in V')
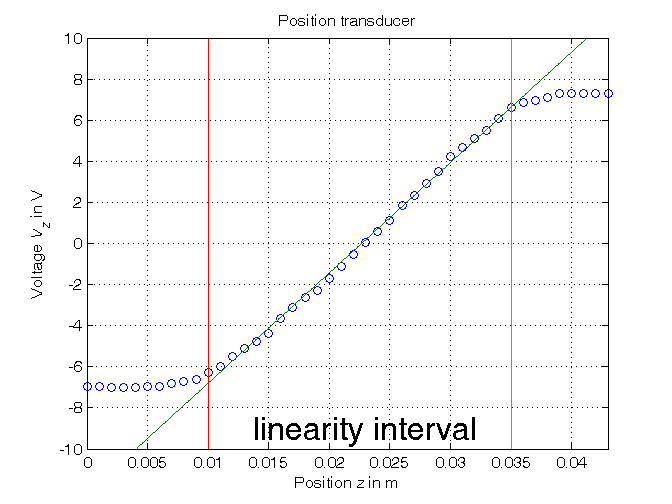
Parametric estimation of a linear model (w.r.t. data) using least squares
% Step3: computation of the parameter estimate % definition of the linearity interval of the characteristic i1=11; % z(11) = 0.01 i2=36; % z(36) = 0.035 z_lin=z(i1:i2); Vz_lin=Vz(i1:i2); N_lin=length(z_lin); % parameter estimation by means of least squares algorithm Phi=[Vz_lin, ones(N_lin,1)]; p=Phi\z_lin; % Form #1: using the "\" operator (more realiable) Kt=1/p(1) Vo=-p(2)/p(1) p_=inv(Phi'*Phi)*Phi'*z_lin; % Form #2: using the pseudoinverse matrix Kt_=1/p_(1) Vo_=-p_(2)/p_(1) % Step 4: graphical comparison of the results Vz0=linspace(-10,10,1000); z_hat=p(1)*Vz0+p(2); figure, plot(z,Vz,'o', z_hat,Vz0,'-', ... z_lin(1)*[1,1],[-10,10],'-', z_lin(end)*[1,1],[-10,10],'-'), axis([min(z),max(z),-10,10]), grid, title('Position transducer'), xlabel('Position{\it z} in m'), ylabel('Voltage{\it V_z} in V'), text(0.013,-9,'\fontsize{20} linearity interval')
Kt = 537.0036 Vo = -12.1779 Kt_ = 537.0036 Vo_ = -12.1779
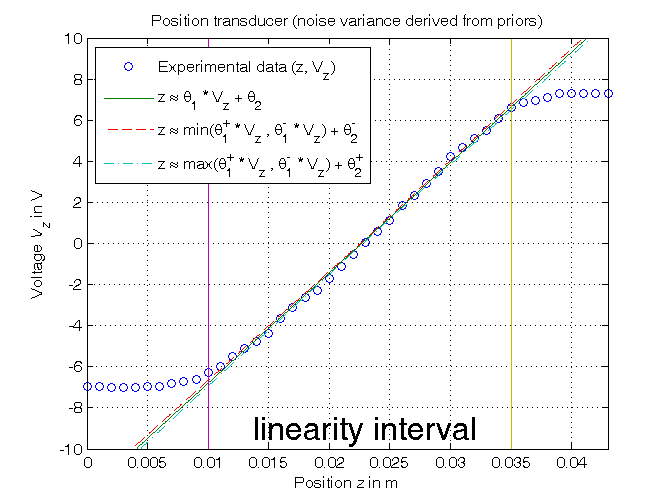
Computation of the parameter confidence intervals (noise variance derived from priors)
% Step 5: computation of the confidence intervals % Definition of the x% confidence intervals: % x=95.4 => k=2 ("2 sigma"); x=99.7 => k=3 ("3 sigma") k_e=2; k_p=2; noise_max=5e-4; sigma_e=noise_max/k_e Sigma_e=sigma_e^2*eye(N_lin); Sigma_p=inv(Phi'*inv(Sigma_e)*Phi); sigma_p=sqrt(diag(Sigma_p)); delta_p=k_p*sigma_p; p_min=p-delta_p; p_max=p+delta_p; Kt_min=1/p_max(1) Kt_max=1/p_min(1) Vo_min=-p_max(2)/p_min(1) Vo_max=-p_min(2)/p_max(1) % Step 6: graphical comparison of the results z_min=min([p_max(1)*Vz0; p_min(1)*Vz0])+p_min(2); z_max=max([p_max(1)*Vz0; p_min(1)*Vz0])+p_max(2); figure, plot(z,Vz,'o', z_hat,Vz0,'-', ... z_min,Vz0,'--', z_max,Vz0,'-.', ... z_lin(1)*[1,1],[-10,10],'-',z_lin(end)*[1,1],[-10,10],'-') axis([min(z),max(z),-10,10]), grid, title('Position transducer (noise variance derived from priors)'), xlabel('Position{\it z} in m'), ylabel('Voltage{\it V_z} in V'), legend('Experimental data (z, V_z)_{ }^{ }', ... 'z \approx \theta_{1} * V_{z} + \theta_2', ... 'z \approx min(\theta_{1}^{+} * V_{z} , \theta_{1}^{-} * V_{z}) + \theta_{2}^{-}', ... 'z \approx max(\theta_{1}^{+} * V_{z} , \theta_{1}^{-} * V_{z}) + \theta_{2}^{+}', 2), text(0.013,-9,'\fontsize{20} linearity interval')
sigma_e = 2.5000e-04 Kt_min = 530.0654 Kt_max = 544.1257 Vo_min = -12.3928 Vo_max = -11.9685
Computation of the parameter confidence intervals (noise variance estimated from data)
% Step 7: computation of the confidence intervals sigma_e_ml=sqrt((z_lin-Phi*p)'*(z_lin-Phi*p)/N_lin) Sigma_e=sigma_e_ml^2*eye(N_lin); Sigma_p=inv(Phi'*inv(Sigma_e)*Phi); sigma_p=sqrt(diag(Sigma_p)); delta_p=k_p*sigma_p; p_min=p-delta_p; p_max=p+delta_p; Kt_min=1/p_max(1) Kt_max=1/p_min(1) Vo_min=-p_max(2)/p_min(1) Vo_max=-p_min(2)/p_max(1) % Step 8: graphical comparison of the results z_min=min([p_max(1)*Vz0; p_min(1)*Vz0])+p_min(2); z_max=max([p_max(1)*Vz0; p_min(1)*Vz0])+p_max(2); figure, plot(z,Vz,'o', z_hat,Vz0,'-', ... z_min,Vz0,'--', z_max,Vz0,'-.', ... z_lin(1)*[1,1],[-10,10],'-',z_lin(end)*[1,1],[-10,10],'-') axis([min(z),max(z),-10,10]), grid, title('Position transducer (noise variance estimated from data)'), xlabel('Position{\it z} in m'), ylabel('Voltage{\it V_z} in V'), legend('Experimental data (z, V_z)_{ }^{ }', ... 'z \approx \theta_{1} * V_{z} + \theta_2', ... 'z \approx min(\theta_{1}^{+} * V_{z} , \theta_{1}^{-} * V_{z}) + \theta_{2}^{-}', ... 'z \approx max(\theta_{1}^{+} * V_{z} , \theta_{1}^{-} * V_{z}) + \theta_{2}^{+}', 2), text(0.013,-9,'\fontsize{20} linearity interval')
sigma_e_ml = 3.5618e-04 Kt_min = 527.1726 Kt_max = 547.2082 Vo_min = -12.4858 Vo_max = -11.8813

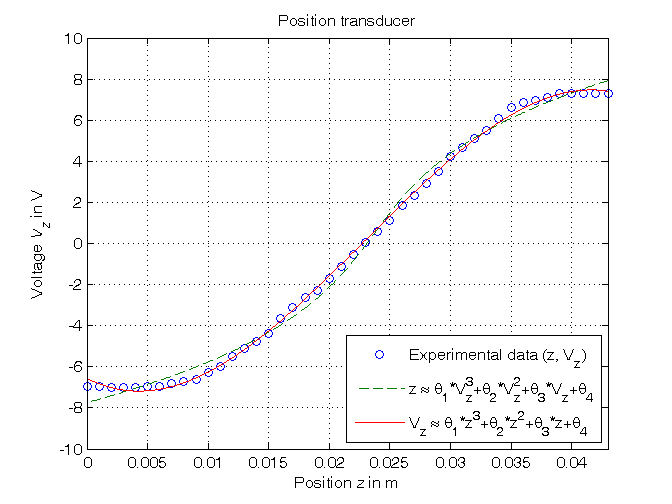
Parametric estimation of 3rd order polynomial models (w.r.t. data) using least squares
% Step 9: computation of the parameter estimate % a) assuming Vz as independent variable, z as variable dependent on Vz => % z = theta(1)*Vz^3 + theta(2)*Vz^2 + theta(3)*Vz + theta(4) Phi=[Vz.^3, Vz.^2, Vz, Vz.^0]; format shortE, format compact p3a=Phi\z % Form #1: using the "\" operator z_pol=polyval(p3a,Vz0); p3a_=polyfit(Vz,z,3) % Form #2: using the "polyfit" function format short, format compact % b) assuming z as independent variable, Vz as variable dependent on z => % Vz = theta(1)*z^3 + theta(2)*z^2 + theta(3)*z + theta(4) Phi=[z.^3, z.^2, z, z.^0]; p3b=Phi\Vz % Form #1: using the "\" operator z0=linspace(min(z),max(z),1000); Vz_pol=polyval(p3b,z0); p3b_=polyfit(z,Vz,3) % Form #2: using the "polyfit" function % Step 10: graphical comparison of the results figure, plot(z,Vz,'o',z_pol,Vz0,'--',z0,Vz_pol,'-'), axis([min(z),max(z),-10,10]), grid, title('Position transducer'), xlabel('Position{\it z} in m'), ylabel('Voltage{\it V_z} in V'), legend('Experimental data (z, V_z)_{ }^{ }', ... 'z \approx \theta_1*V_z^3+\theta_2*V_z^2+\theta_3*V_z+\theta_4', ... 'V_z \approx \theta_1*z^3+\theta_2*z^2+\theta_3*z+\theta_4',4)
p3a =
2.4097e-05
-3.4590e-05
1.2715e-03
2.3100e-02
p3a_ =
2.4097e-05 -3.4590e-05 1.2715e-03 2.3100e-02
p3b =
1.0e+05 *
-5.6269
0.3877
-0.0030
-0.0001
p3b_ =
1.0e+05 *
-5.6269 0.3877 -0.0030 -0.0001